Analyzing MiniSEED seismic data in MATLAB (codes included)
We will learn how to convert a mseed data file into mat format and then read and analyze it using MATLAB
I have a MiniSEED (or mseed) file and I want to analyze it in MATLAB. But unfortunately, MATLAB can’t read mseed file. So, let us figure out how can we read and analyze it using MATLAB.
Similar posts
What is MiniSEED format?
IRIS uses SEED as a data format intended primarily for the archival and exchange of seismological time series data and related metadata. MiniSEED is a stripped down version of SEED containing only waveform data. There is no station and channel metadata included. See here for more.
Utitlity program to convert MiniSEED into MAT format
I wrote a utility program that uses the Obspy library in Python to convert the mseed file to mat format. You can download the utility from here.
usage: convert_mseed_mat.py [-h] -inp INPUT_MSEED [-out OUTPUT_MAT]
Python utility program to convert mseed file to mat (by Utpal Kumar, IESAS, 2021/04)
optional arguments:
-h, --help show this help message and exit
-inp INPUT_MSEED, --input_mseed INPUT_MSEED
input mseed file, e.g. example_2020-05-01_IN.RAGD..BHZ.mseed
-out OUTPUT_MAT, --output_mat OUTPUT_MAT
output mat file name, e.g. example_2020-05-01_IN.RAGD..BHZ.mat
Let us see an example:
python convert_mseed_mat.py -inp example_2020-05-01_IN.RAGD..BHZ.mseed
Output data structure
statscontains all the meta data information corresponding to each trace anddatacontain the time series data
mat_file.mat -> stats, data
stats -> stats_0, stats_1, ...
data -> data_0, data_1, ...
Read mat file in MATLAB
Now, let us read the mat file containing the seismic time series data. We start by the usual initializing the MATLAB and reading the file name.
clear; close all; clc;
wdir='.\';
fileloc0=[wdir,'example_2020-05-01_IN.RAGD..BHZ'];
fileloc_ext = '.mat';
fileloc = [fileloc0 fileloc_ext];
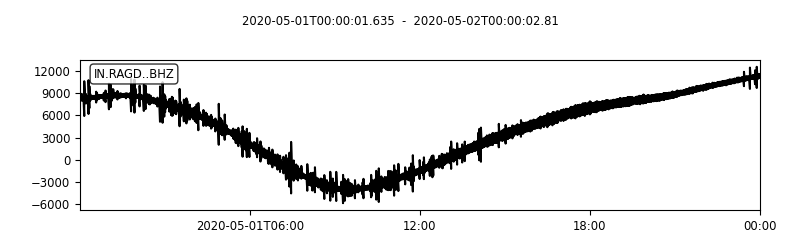
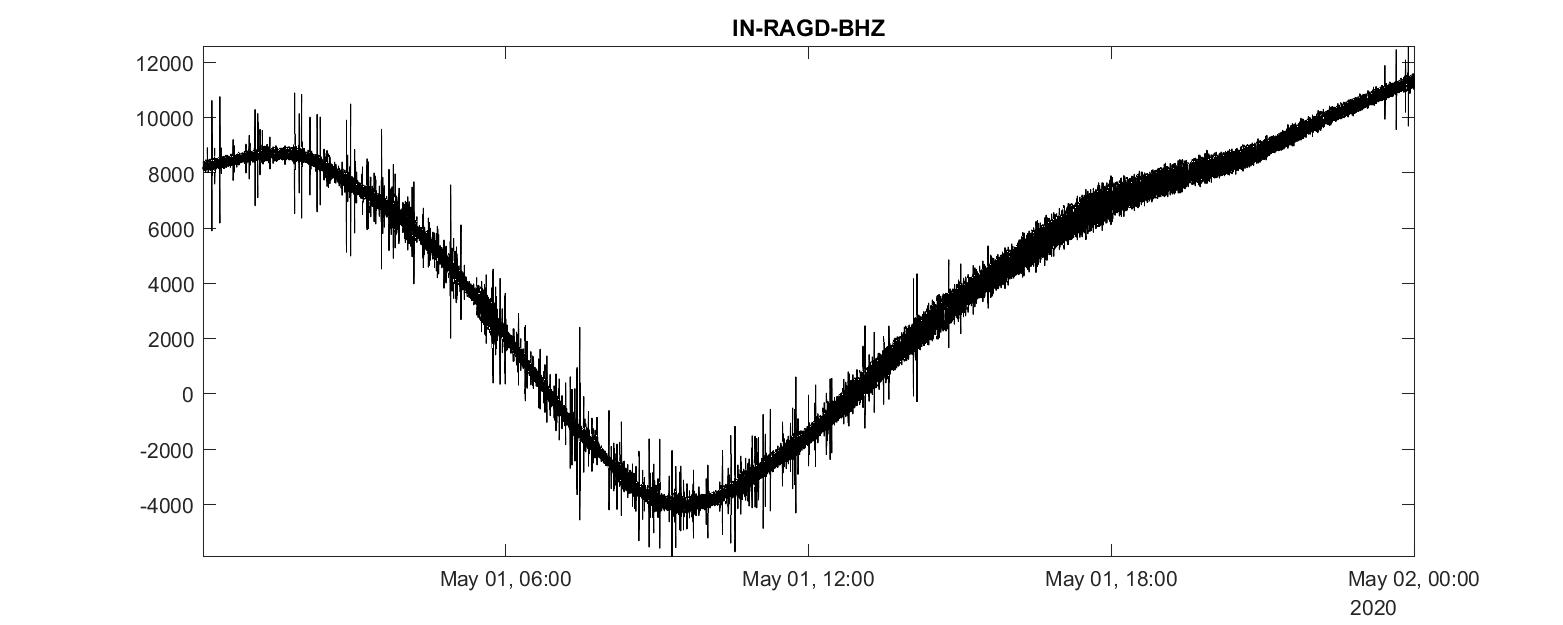
Plot time series
We now check if the mat file exists, and the read the meta data stored in stats_0. We get the sampling_rate, delta, starttime, endtime. For plotting, we create the datetime_array.
if exist(fileloc,'file')
disp(['File exists ', fileloc]);
load(fileloc);
all_stats = fieldnames(stats);
all_data = fieldnames(data);
% for id=1:length(fieldnames(data))
for id=1
stats_0 = stats.(all_stats{id});
data_0 = data.(all_data{id});
sampling_rate = getfield(stats_0,'sampling_rate');
delta = getfield(stats_0,'delta');
starttime = getfield(stats_0,'starttime');
endtime = getfield(stats_0,'endtime');
t1 = datetime(starttime,'InputFormat',"yyyy-MM-dd'T'HH:mm:ss.SSS'Z'");
t2 = datetime(endtime,'InputFormat',"yyyy-MM-dd'T'HH:mm:ss.SSS'Z'");
datetime_array = t1:seconds(delta):t2;
%% plot time series
fig = figure('Renderer', 'painters', 'Position', [100 100 1000 400], 'color','w');
plot(t1:seconds(delta):t2, data_0, 'k-')
title([getfield(stats_0,'network'),'-', getfield(stats_0,'station'), '-', getfield(stats_0,'channel')])
axis tight;
print(fig,['docs/',fileloc0, '_ts', num2str(id),'.jpg'],'-djpeg')
% close all;
end
end
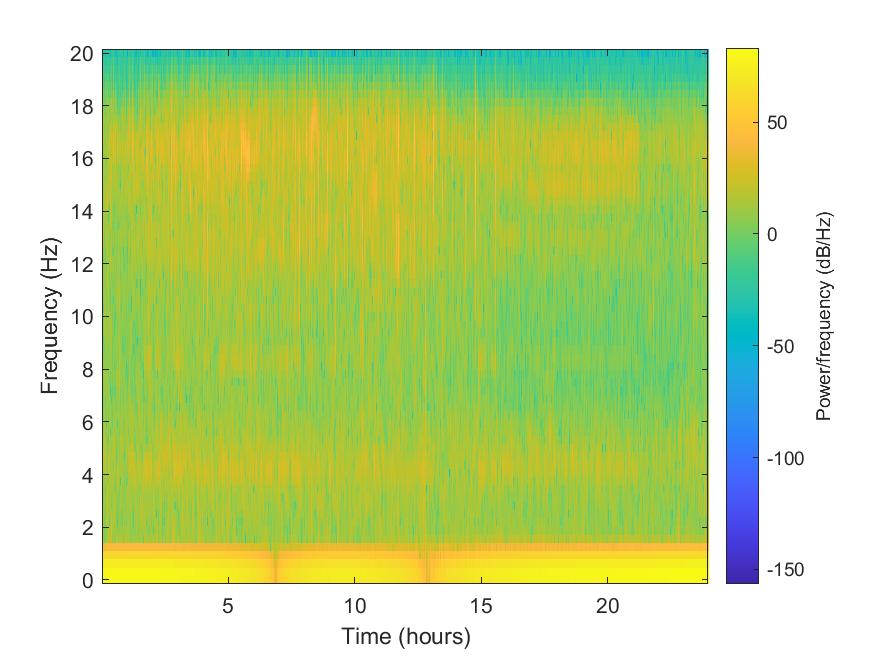
Plot spectrogram
We used the spectrogram function from MATLAB to plot the spectrogram (can be improved further). We divide the signal into sections of length 128, windowed with a Kaiser window with shape parameter (\beta = 18) and specify 120 samples of overlap between adjoining sections. We evaluate the spectrum at 65 frequencies and ((length(x)−120)/(128−120)=235) time bins.
if exist(fileloc,'file')
disp(['File exists ', fileloc]);
load(fileloc);
all_stats = fieldnames(stats);
all_data = fieldnames(data);
% for id=1:length(fieldnames(data))
for id=1
stats_0 = stats.(all_stats{id});
data_0 = data.(all_data{id});
sampling_rate = getfield(stats_0,'sampling_rate');
fig2 = figure('Renderer', 'painters', 'Position', [100 100 1000 400], 'color','w');
data_0_double = double(data_0);
spectrogram(data_0_double,kaiser(128,18),120,128,sampling_rate,'yaxis')
%% if you want to normalize the frequency axis in range 0 to 1
% yticks([0 sampling_rate/4 sampling_rate/2])
% yticklabels({'0','0.5','1'})
% ylabel('Normalized Frequency');
title([getfield(stats_0,'network'),'-', getfield(stats_0,'station'), '-', getfield(stats_0,'channel')])
print(fig2,['docs/',fileloc0, '_spectrogram', num2str(id),'.jpg'],'-djpeg')
% close all;
end
end
Conclusions
Converting Miniseed into mat format allows us to easily read the seismic time series data in MATLAB. Once we load the data in MATLAB, we can make use of all the avilable MATLAB commands and tools.
Disclaimer of liability
The information provided by the Earth Inversion is made available for educational purposes only.
Whilst we endeavor to keep the information up-to-date and correct. Earth Inversion makes no representations or warranties of any kind, express or implied about the completeness, accuracy, reliability, suitability or availability with respect to the website or the information, products, services or related graphics content on the website for any purpose.
UNDER NO CIRCUMSTANCE SHALL WE HAVE ANY LIABILITY TO YOU FOR ANY LOSS OR DAMAGE OF ANY KIND INCURRED AS A RESULT OF THE USE OF THE SITE OR RELIANCE ON ANY INFORMATION PROVIDED ON THE SITE. ANY RELIANCE YOU PLACED ON SUCH MATERIAL IS THEREFORE STRICTLY AT YOUR OWN RISK.

Leave a comment